Tutorial
Decorators
@timeit_arg_info_dec
New in v0.1.0
timeit_arg_info_dec is a decorator that decorates a function when it runs, by printing its used parameters, their arguments, the execution time and the output of that function. This can help e.g. with debugging.
[2]:
from typing import List
from time import sleep
import pandas as pd
from extra_ds_tools.decorators.func_decorators import timeit_arg_info_dec
@timeit_arg_info_dec(round_seconds=1)
def illustrate_decorater(a_number: int,
text: str,
lst: List[int],
df: pd.DataFrame,
either: bool = True,
*args,
**kwargs):
sleep(1)
return "Look how informative!"
illustrate_decorater(42, 'Bob', list(range(100)), pd.DataFrame([list(range(1,10))]), either=False, **{'Even': 'this works!'})
illustrate_decorater()
---------------------------------------------------------------------------------------------------------------------------------
param type_hint default_value arg_type arg_value arg_len
-- ------------- --------------------------- --------------- --------------------------- ------------------------ ---------
0 a_number int int 42
1 text str str Bob 3
2 lst List[int] list [0, 1, 2, .. 7, 98, 99] 100
3 df pandas.core.frame.DataFrame pandas.core.frame.DataFrame (1, 9)
4 either bool True bool False
5 kwarg['Even'] str this works! 11
illustrate_decorater()took 1.0 seconds to run.
Returned:
Look how informative!
---------------------------------------------------------------------------------------------------------------------------------
[2]:
'Look how informative!'
For the full documentation of timeit_arg_info_dec click here.
Plots
try_diff_distribution_plots
New in v0.2.0
try_diff_distribution_plots is a function which performs different transformations to a list of numerical values and plots the histogram, probability and the boxplot for each transformation.
[1]:
from numpy.random import default_rng
from extra_ds_tools.plots.eda import try_diff_distribution_plots
rng = default_rng(42)
values = rng.pareto(a=100, size=1000)
fig, axes, transformed_values = try_diff_distribution_plots(values, hist_bins=40)
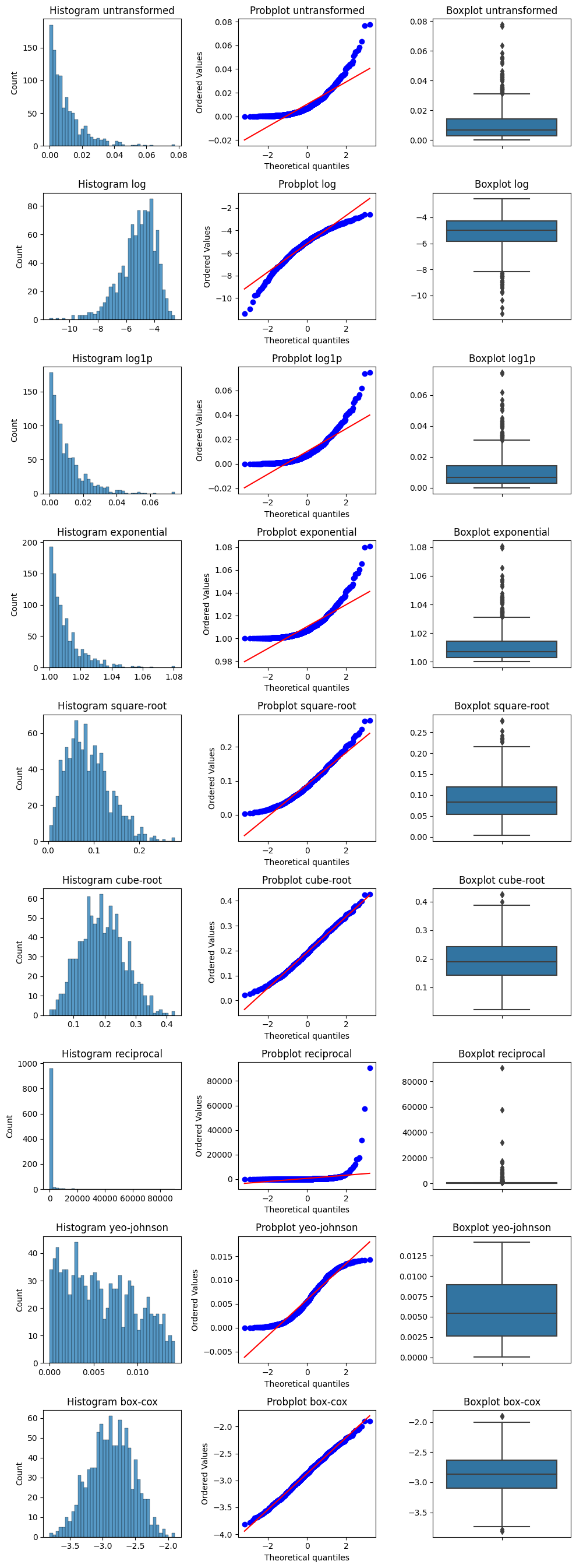
For the full documentation of try_diff_distribution_plots click here.
stripboxplot
New in v0.3.0
stripboxplot is a plot which combines seaborn’s boxplot and stripplot into one plot and adds extra count information using extra-datascience-tool’s add_counts_to_yticks and add_counts_to_xticks.
[4]:
# import libraries
import pandas as pd
import numpy as np
from extra_ds_tools.plots.eda import stripboxplot
from numpy.random import default_rng
# generate data
rng = default_rng(42)
cats = ['Cheetah', 'Leopard', 'Puma']
cats = rng.choice(cats, size=1000)
cats = np.append(cats, [None]*102)
weights = rng.integers(25, 100, size=1000)
weights = np.append(weights, [np.nan]*100)
weights = np.append(weights, np.array([125,135]))
rng.shuffle(cats)
rng.shuffle(weights)
df = pd.DataFrame({'cats': cats, 'weights': weights})
df.head()
[4]:
| cats | weights | |
|---|---|---|
| 0 | Cheetah | 86.0 |
| 1 | Puma | 38.0 |
| 2 | Puma | 68.0 |
| 3 | None | NaN |
| 4 | Puma | 36.0 |
[5]:
# run stripboxplot
fig, ax = stripboxplot(df, 'cats', 'weights')
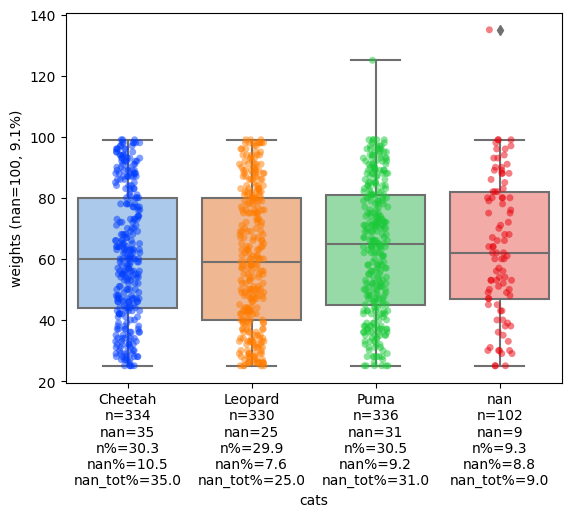
For the full documentation of stripboxplot click here.
add_counts_to_xticks
New in v0.3.0
add_counts_to_xticks is a function which add count statistics of a categorical variable on the x-axis of a plot. If the categorical variable is on the y-axis you can use add_counts_to_yticks instead.
[1]:
# import libraries
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
import numpy as np
from extra_ds_tools.plots.format import add_counts_to_xticks
from numpy.random import default_rng
# generate data
rng = default_rng(42)
cats = ['Cheetah', 'Leopard', 'Puma']
cats = rng.choice(cats, size=1000)
cats = np.append(cats, [None]*102)
weights = rng.integers(25, 100, size=1000)
weights = np.append(weights, [np.nan]*100)
weights = np.append(weights, np.array([125,135]))
rng.shuffle(cats)
rng.shuffle(weights)
df = pd.DataFrame({'cats': cats, 'weights': weights})
df.head()
[1]:
| cats | weights | |
|---|---|---|
| 0 | Cheetah | 86.0 |
| 1 | Puma | 38.0 |
| 2 | Puma | 68.0 |
| 3 | None | NaN |
| 4 | Puma | 36.0 |
Create e.g. a violinplot
[2]:
fig, ax = plt.subplots()
sns.violinplot(df, x='cats', y='weights', ax=ax)
[2]:
<AxesSubplot: xlabel='cats', ylabel='weights'>
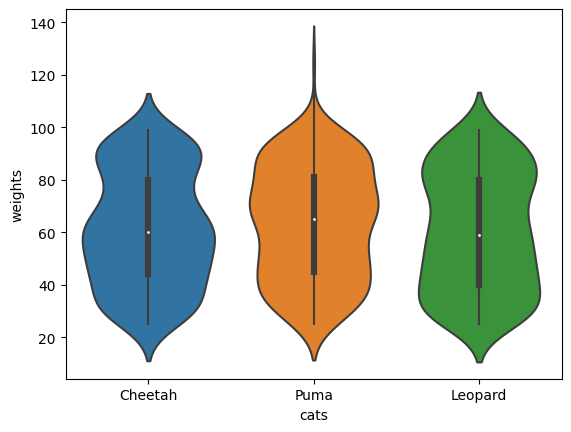
Add counts to the x-ticks
[3]:
fig, ax = add_counts_to_xticks(fig, ax, df, x_col='cats', y_col='weights')
fig
[3]:
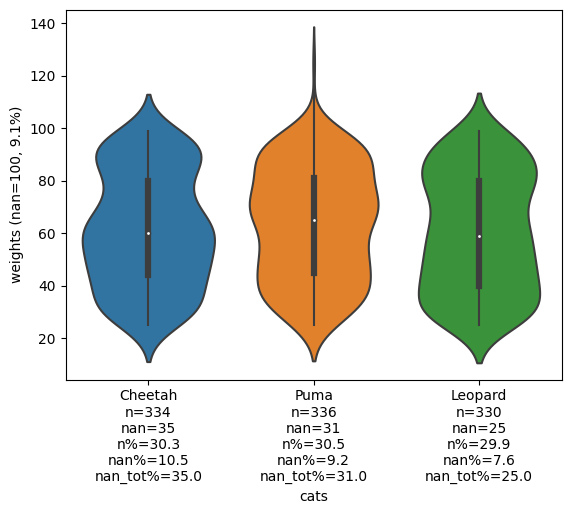
For the full documentation of add_counts_to_xticks click here.
ML
filter_tried_params
New in v0.4.0
filter_tried_params is a function which filters out previously tried parameters in a GridSearchCV if the model is otherwise identical. This can save a lot of time because you won’t be rerunning already tried settings.
[1]:
# import libraries
from sklearn.model_selection import GridSearchCV
from sklearn.tree import DecisionTreeRegressor
from sklearn.pipeline import make_pipeline
from extra_ds_tools.ml.sklearn.model_selection import filter_tried_params
model = make_pipeline(DecisionTreeRegressor())
new_param_grid = {
"decisiontreeregressor__max_depth": [1, 2],
"decisiontreeregressor__splitter": ["best", "random"],
}
new_gridsearch = GridSearchCV(model, new_param_grid)
# initiate two other GridsearchCVs, we assume we have ran them already
tried_param_grid1 = {
"decisiontreeregressor__max_depth": [2, 3],
"decisiontreeregressor__splitter": ["best", "random"],
}
tried_param_grid2 = {
"decisiontreeregressor__max_depth": [3, 4],
"decisiontreeregressor__splitter": ["best", "random"],
}
tried_gridsearches = [
GridSearchCV(model, tried_param_grid1),
GridSearchCV(model, tried_param_grid2)
]
# change the param grid of the new GridSearchCV to the filtered param grid
untried_param_grid = filter_tried_params(gridsearchcv=new_gridsearch, tried_gridsearches=tried_gridsearches)
new_gridsearch.param_grid = untried_param_grid
new_gridsearch.param_grid
[1]:
[{'decisiontreeregressor__max_depth': [1],
'decisiontreeregressor__splitter': ['best']},
{'decisiontreeregressor__max_depth': [1],
'decisiontreeregressor__splitter': ['random']}]
As you can see above, the new GridSearchCV will only run the two new options it hasn’t tried before.
For the full documentation of filter_tried_params click here.
EstimatorSwitch
New in v0.5.0
EstimatorSwitch is a meta-estimator that can turn on or off other estimators/transformers in a scikit-learn Pipeline. This can e.g. be useful when testing for the impact of a feature-engineering estimator/transformer with a GridSearchCV.
[1]:
# import libraries
import numpy as np
from sklearn.pipeline import Pipeline
from feature_engine.imputation import MeanMedianImputer, ArbitraryNumberImputer
from extra_ds_tools.ml.sklearn.meta_estimators import EstimatorSwitch
# create X and y
X = np.array([np.nan, 10] * 5).reshape(-1,1)
y = np.array([5, 10] * 5).reshape(-1, 1)
# create pipeline steps
pipeline = Pipeline(
[("medianimputer", EstimatorSwitch(MeanMedianImputer(), apply=False)),
("arbitraryimputer", EstimatorSwitch(ArbitraryNumberImputer(-1), apply=True))]
)
# transform X according to pipeline
pipeline.fit_transform(X)
[1]:
| x0 | |
|---|---|
| 0 | -1.0 |
| 1 | 10.0 |
| 2 | -1.0 |
| 3 | 10.0 |
| 4 | -1.0 |
| 5 | 10.0 |
| 6 | -1.0 |
| 7 | 10.0 |
| 8 | -1.0 |
| 9 | 10.0 |
Above you can see that the nan values have not been imputed with the mean: 10, but with the arbitrary number -1 because we used EstimatorSwitch to turn off the MeanMedianImputer.
To use EstimatorSwitch to turn off estimators/transformers during a GridSearchCV:
[2]:
import pandas as pd
from sklearn.tree import DecisionTreeRegressor
from sklearn.model_selection import GridSearchCV
from sklearn.pipeline import Pipeline
from feature_engine.imputation import MeanMedianImputer, ArbitraryNumberImputer
from extra_ds_tools.ml.sklearn.meta_estimators import EstimatorSwitch
[3]:
clf = Pipeline(
[("meanmedianimputer", EstimatorSwitch(MeanMedianImputer())),
("arbitraryimputer", EstimatorSwitch(ArbitraryNumberImputer(-1))),
("tree", DecisionTreeRegressor())]
)
clf
[3]:
Pipeline(steps=[('meanmedianimputer',
EstimatorSwitch(estimator=MeanMedianImputer())),
('arbitraryimputer',
EstimatorSwitch(estimator=ArbitraryNumberImputer(arbitrary_number=-1))),
('tree', DecisionTreeRegressor())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('meanmedianimputer',
EstimatorSwitch(estimator=MeanMedianImputer())),
('arbitraryimputer',
EstimatorSwitch(estimator=ArbitraryNumberImputer(arbitrary_number=-1))),
('tree', DecisionTreeRegressor())])EstimatorSwitch(estimator=MeanMedianImputer())
MeanMedianImputer()
MeanMedianImputer()
EstimatorSwitch(estimator=ArbitraryNumberImputer(arbitrary_number=-1))
ArbitraryNumberImputer(arbitrary_number=-1)
ArbitraryNumberImputer(arbitrary_number=-1)
DecisionTreeRegressor()
Use the get_params() method to get the right parameters for the parameter grid for the GridSearchCV.
[4]:
clf.get_params()
[4]:
{'memory': None,
'steps': [('meanmedianimputer',
EstimatorSwitch(estimator=MeanMedianImputer())),
('arbitraryimputer',
EstimatorSwitch(estimator=ArbitraryNumberImputer(arbitrary_number=-1))),
('tree', DecisionTreeRegressor())],
'verbose': False,
'meanmedianimputer': EstimatorSwitch(estimator=MeanMedianImputer()),
'arbitraryimputer': EstimatorSwitch(estimator=ArbitraryNumberImputer(arbitrary_number=-1)),
'tree': DecisionTreeRegressor(),
'meanmedianimputer__apply': True,
'meanmedianimputer__estimator__imputation_method': 'median',
'meanmedianimputer__estimator__variables': None,
'meanmedianimputer__estimator': MeanMedianImputer(),
'arbitraryimputer__apply': True,
'arbitraryimputer__estimator__arbitrary_number': -1,
'arbitraryimputer__estimator__imputer_dict': None,
'arbitraryimputer__estimator__variables': None,
'arbitraryimputer__estimator': ArbitraryNumberImputer(arbitrary_number=-1),
'tree__ccp_alpha': 0.0,
'tree__criterion': 'squared_error',
'tree__max_depth': None,
'tree__max_features': None,
'tree__max_leaf_nodes': None,
'tree__min_impurity_decrease': 0.0,
'tree__min_samples_leaf': 1,
'tree__min_samples_split': 2,
'tree__min_weight_fraction_leaf': 0.0,
'tree__random_state': None,
'tree__splitter': 'best'}
Create a parameter grid with MeanMedianImputer and the ArbitraryNumberImputer turned off and on seperately using the apply parameter of EstimatorSwitch.
[5]:
param_grid = [
{
"meanmedianimputer__apply": [True],
"meanmedianimputer__estimator__imputation_method": ["mean", "median"],
"arbitraryimputer__apply": [False]
},
{
"arbitraryimputer__apply": [True],
"arbitraryimputer__estimator__arbitrary_number": [-1],
"meanmedianimputer__apply": [False]
}
]
[6]:
gridsearch = GridSearchCV(estimator=clf, param_grid=param_grid)
gridsearch.fit(X,y)
pd.DataFrame(gridsearch.cv_results_)
[6]:
| mean_fit_time | std_fit_time | mean_score_time | std_score_time | param_arbitraryimputer__apply | param_meanmedianimputer__apply | param_meanmedianimputer__estimator__imputation_method | param_arbitraryimputer__estimator__arbitrary_number | params | split0_test_score | split1_test_score | split2_test_score | split3_test_score | split4_test_score | mean_test_score | std_test_score | rank_test_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.001864 | 0.000288 | 0.001197 | 0.000127 | False | True | mean | NaN | {'arbitraryimputer__apply': False, 'meanmedian... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2 |
| 1 | 0.001910 | 0.000170 | 0.001023 | 0.000098 | False | True | median | NaN | {'arbitraryimputer__apply': False, 'meanmedian... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2 |
| 2 | 0.001154 | 0.000081 | 0.000985 | 0.000176 | True | False | NaN | -1 | {'arbitraryimputer__apply': True, 'arbitraryim... | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1 |
For the full documentation of EstimatorSwitch click here.